Summary of side projects, end of 2025
Just a small recap of some of the things I worked or am still working on, which are not big enough or not ready to be given a whole post.
AdminSCH DNS adblocker
I already used Pi-hole in my homelab, which, if you didn't know, is a lightweight DNS server with the extra feature of blocklisting. This blocklisting enables users to provide multiple lists of hosts, which will be resolved to 0.0.0.0, essentially blocking whatever it should have loaded. This makes it a useful adblocker, because you just need to load a list of domains of adware, malware etc. sites, and it will block them even before it would reach the client. Of course these domain are changing in the minute so false negatives are out there, and also false positives are a thing too, where useful content is blocked, making services unusable, but this is not usual.
So long story short, I thought why not integrate an adblocking solution into the dormitory network we have at Schönherz. This was a software development work, where I worked on the AdminSCH application which is the service for the college students, where they need to register their devices MAC address, which enables them to connect to the internet. AdminSCH is made out of many microservices, each responsible for a discrete functionality. For adblocking to work, a Pi-hole DNS server was deployed in the dormitory infrastructure as k8s service. The high level flow, is that the user registers a device, where she can also opt-in for this DNS adblocking, the microservice syncs with the DHCP server, which will serve the IP address with the standard DNS server address, or the Pi-hole address, depending on the adblocking boolean the user given on the frontend. The registration model schema was expanded to store this boolean, and the logic was added to the DHCP server, to sever different DNS sever addresses.
A small snippet of the frontend, where a user can manage her registrations, including the DNS adblocking:
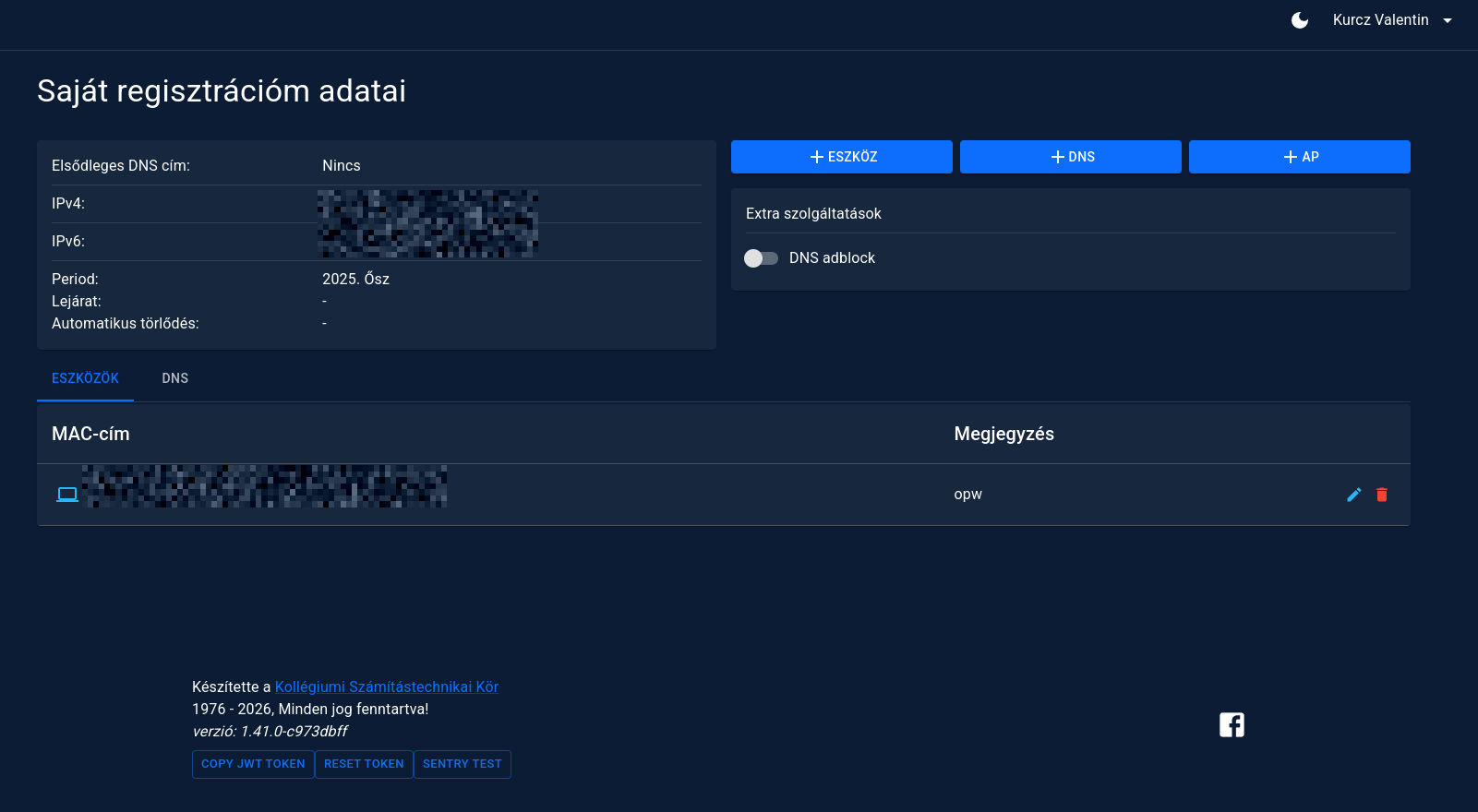
I decided this feature should be opt-in, because of the possibility of false positives.
Currently we are blocking approximately 288.000 domains, and as a result, 20% of the queries are resolved to 0.0.0.0. Crazy to think that even with a permissive list like 288 thousand, the 20% of the traffic is just companies trying to track us with third-party cookies, or trying to sell something.
Oh and as a little side effect, this also blocks known malware distributing domains, protecting students even more. Obviously, this is a cat and mouse game, so there is no guarantee, that no malware will reach the end users, but at least it's another safety net.
Malware clustering and static analysis
Utilizing my free time between work and 2 semesters, I started to read books about malware analysis. I had prior experience in reverse engineering from CTFs, but never ever had the means to start malware analysis. I'm currently reading these two books in this topic:-sikorski
-
Malware Data Science - Attack Detection and Attribution by Joshua Saxe with Hillary Sanders

-
Practical Malware Analysis - Michael Sikorski and Andrew Honig

These two books fueled my interest so much, that I decided that I will continue my work at my university in this trajectory.
But in the mean time, I started a little pilot, to practice some skills and put the knowledge from the malware data science book to use. This application would enable users to upload binary samples, or search already uploaded samples by hash. Many type of analysis could be done on the samples:
- Extracting features, like Import Address Table from PE binaries, strings, headers, N-grams
- Based on these features a Jaccard similarity index can be calculated, revealing if an uploaded sample is similar to anything already present in the database. This is good for clustering malwares into families, and save the effort of manual analysis. Calculating a Jaccard index, which is basically the similarity of two sets, is two expensive, so I rather use MinHashing, which with enough permutations, will converge to the Jaccard index with an acceptable level of error. After MinHashing only the minhashes have to be compared. However, with massive amounts of samples, this linear operation is costly too, so as a remedy I use an LSH index, so minhash comparision will only happen between samples, which is probably more similar than a given threshold, and this will be a sublinear comparision.
- Later on a simple machine learning model, like a random forest, could be trained, to give the user a prediction, if the sample is a malware or not, based on the extracted features.
Currently, I have done the similarity backend in python, connecting to a postgresql database, for storing the sample minhashes, and connecting to a redis key store, to store the LSH index.
import os
import redis
from datasketch import MinHash
from datasketch import MinHashLSH
from typing import List, Tuple
from hashlib import sha1
from sqlalchemy.orm import Session
from sqlalchemy import select, func
from fastapi import FastAPI, UploadFile, Depends, HTTPException
from database import engine, get_db
from models import Base, Sample
from fastapi.middleware.cors import CORSMiddleware
from fastapi.concurrency import run_in_threadpool
from schema import SampleResponse, CountResponse, SearchResponse
Base.metadata.create_all(bind=engine)
REDIS_HOST = os.getenv("REDIS_HOST", "localhost")
REDIS_PORT = int(os.getenv("REDIS_PORT", 6379))
SVELTE_HOST = os.getenv("REDIS_HOST", "localhost")
SVELTE_PORT = int(os.getenv("REDIS_PORT", 5173))
app = FastAPI()
app.add_middleware(
CORSMiddleware,
allow_origins=[f"http://{SVELTE_HOST}:{SVELTE_PORT}"],
allow_credentials=True,
allow_methods=["*"],
allow_headers=["*"],
)
r = redis.Redis(host=REDIS_HOST, port=REDIS_PORT, db=0)
lsh = MinHashLSH(
threshold=0.5,
num_perm=128,
storage_config={
'type': 'redis',
'basename': b'sample_lsh',
'redis': {
'host': REDIS_HOST,
'port': REDIS_PORT,
}
}
)
def compute_minhash(content: bytes, perm: int = 128) -> MinHash:
m = MinHash(perm)
for ngram in extract_ngrams(content):
m.update(ngram)
return m
def extract_ngrams(data: bytes, n: int = 4) -> List[bytes]:
if len(data) < n:
return [data]
return [data[i:i+n] for i in range(len(data) - n + 1)]
@app.post("/upload/", response_model=SampleResponse)
async def upload_sample(file: UploadFile, db: Session = Depends(get_db)):
buff_size = 65655
m = sha1()
while (data := await file.read(buff_size)):
m.update(data)
sample_hash = m.hexdigest()
query = select(Sample).where(Sample.hash == sample_hash)
sample = await run_in_threadpool(db.execute(query).scalars().first)
if sample:
return {"message": f"Sample with the hash {sample_hash} already exist in the database", "sample_hash": sample_hash}
else:
await file.seek(0)
minhash = await run_in_threadpool(compute_minhash, await file.read())
sample = Sample(
hash=sample_hash,
minhash=minhash
)
def insert_to_db():
db.add(sample)
db.commit()
await run_in_threadpool(insert_to_db)
lsh.insert(sample.hash, sample.minhash)
if r.exists("sample_count"):
r.incr("sample_count")
return {"message": f"Sample with the hash {sample_hash} added to the database" ,"sample_hash": sample_hash}
@app.get("/samples/count", response_model=CountResponse)
async def sample_count(db: Session = Depends(get_db)):
cached_count = r.get("sample_count")
if cached_count:
return {"count": int(cached_count)}
actual_count = db.scalar(select(func.count()).select_from(Sample))
r.set("sample_count", actual_count, ex=60)
return {"count": actual_count}
@app.get("/search/{sample_hash}", response_model=SearchResponse)
def search(sample_hash: str, db: Session = Depends(get_db)):
query = select(Sample).where(Sample.hash == sample_hash)
if (sample := db.execute(query).scalars().first()):
neighbour_keys = lsh.query(sample.minhash)
neighbor_query = select(Sample).where(Sample.hash.in_(neighbour_keys), Sample.hash != sample.hash)
neighbors = db.execute(neighbor_query).scalars()
results = []
for neighbor in neighbors:
similarity = sample.minhash.jaccard(neighbor.minhash)
results.append({"target_hash": neighbor.hash, "similarity": similarity})
return {"matches": results}
else:
return {"matches": []}
As you can see, only byte ngrams are currently exported from the samples, which is not ideal, and this projects is nowhere near ready. I tried to save as much operation as possible, like for the sample count, I cache that in redis. Also for the IO heavy operations, I run those in a threadpool, not blocking the main thread.
The fronted looks pretty ugly but it gives a frame for the whole application:
I still don't know what will be the end of this, because the scale of this project is bigger than me. I will probably involve others to work on this.